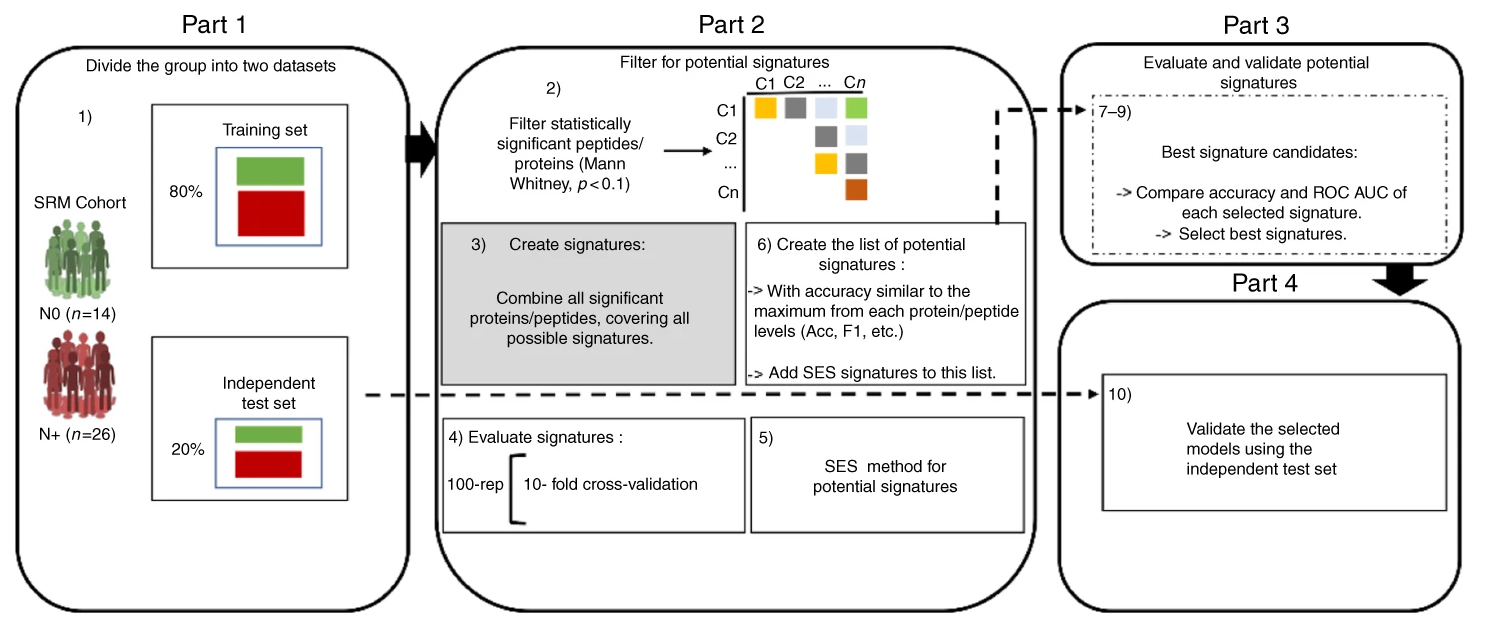
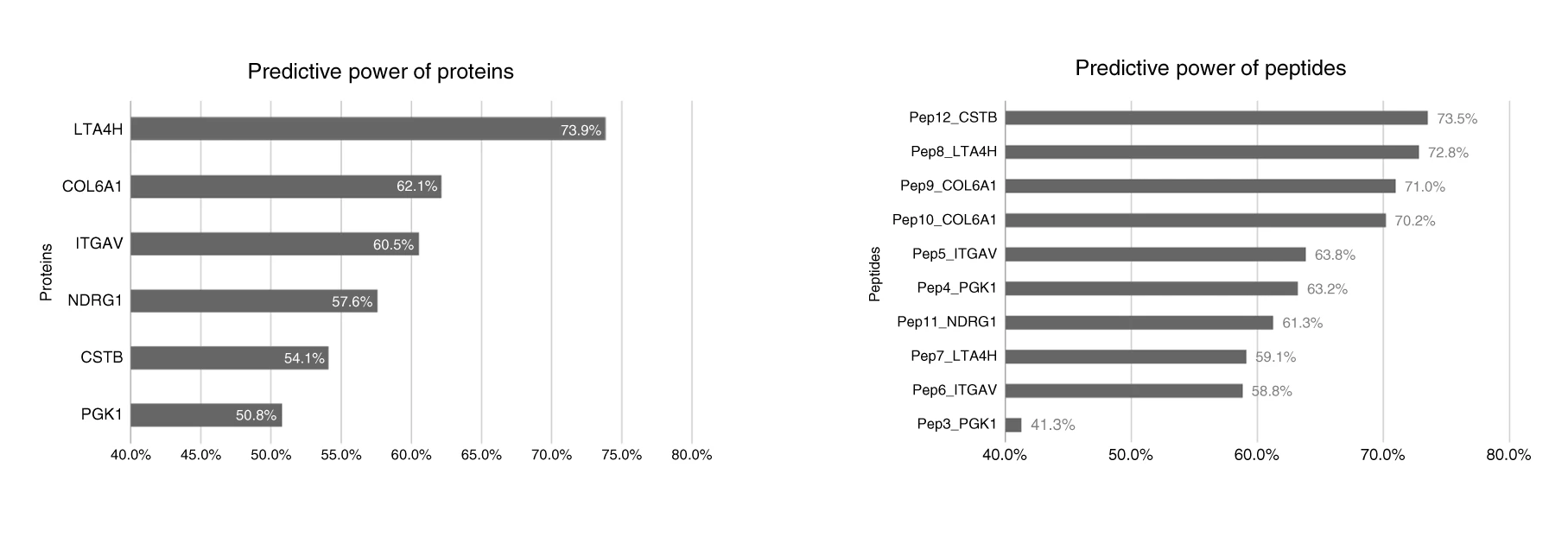
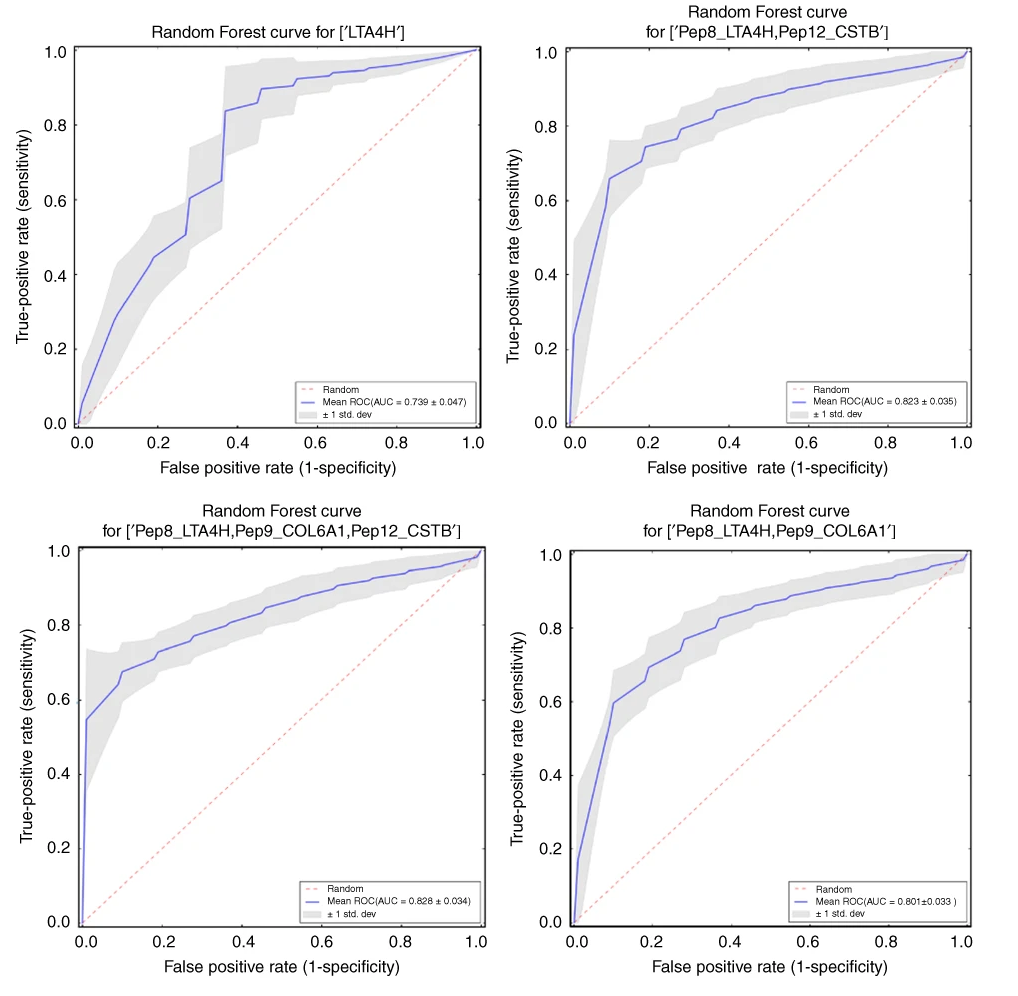
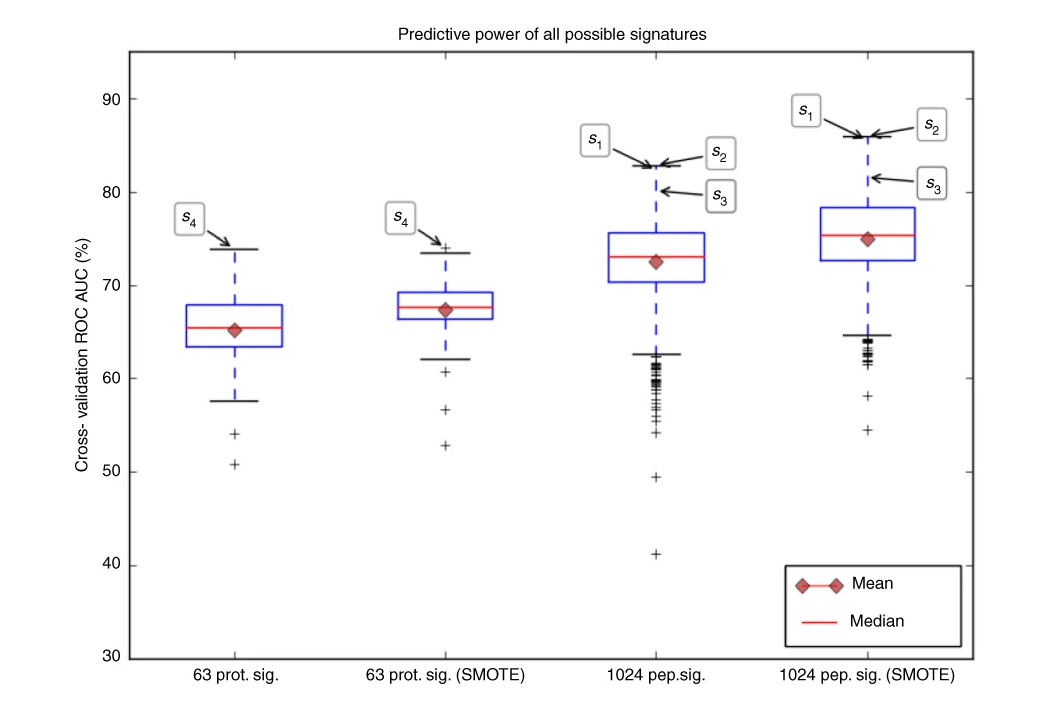
In this project we evaluated proteins candidates for oral cancer biormarkers. Among many analysis and approaches involving discovery and targeted proteomics, in collaboration with the Brazilian Bioscience National Laboratory, I applied statistical and machine learning tools to evaluate the power of potential proteins and peptides candidates signatures to discriminate the N0/N+ oral cancer conditions explained in the article.
Our work was published in Nature Communications and appeared on radio, tv and magazines, and received an award for innovation integrating Computer Science methods and Biology for Health.