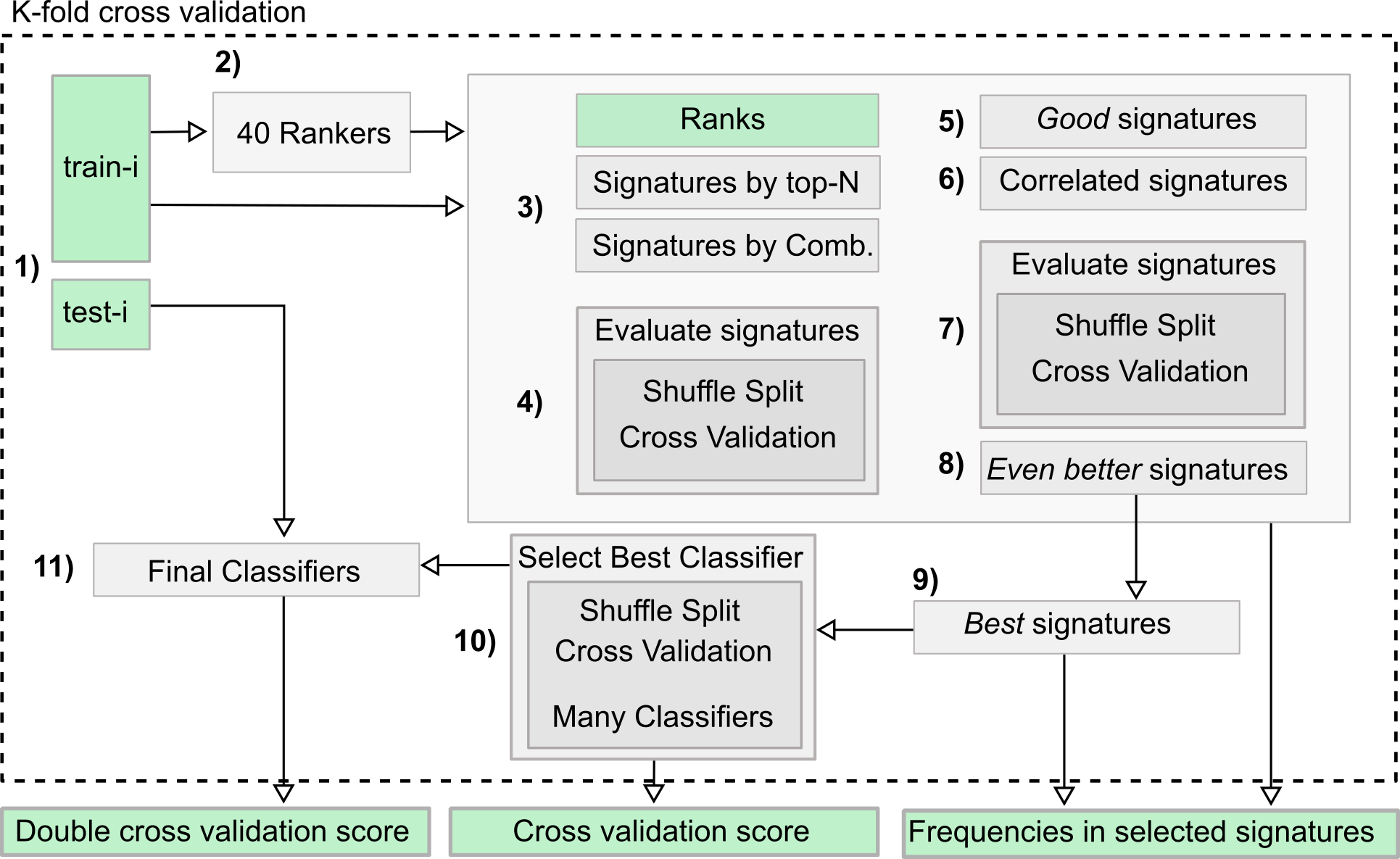
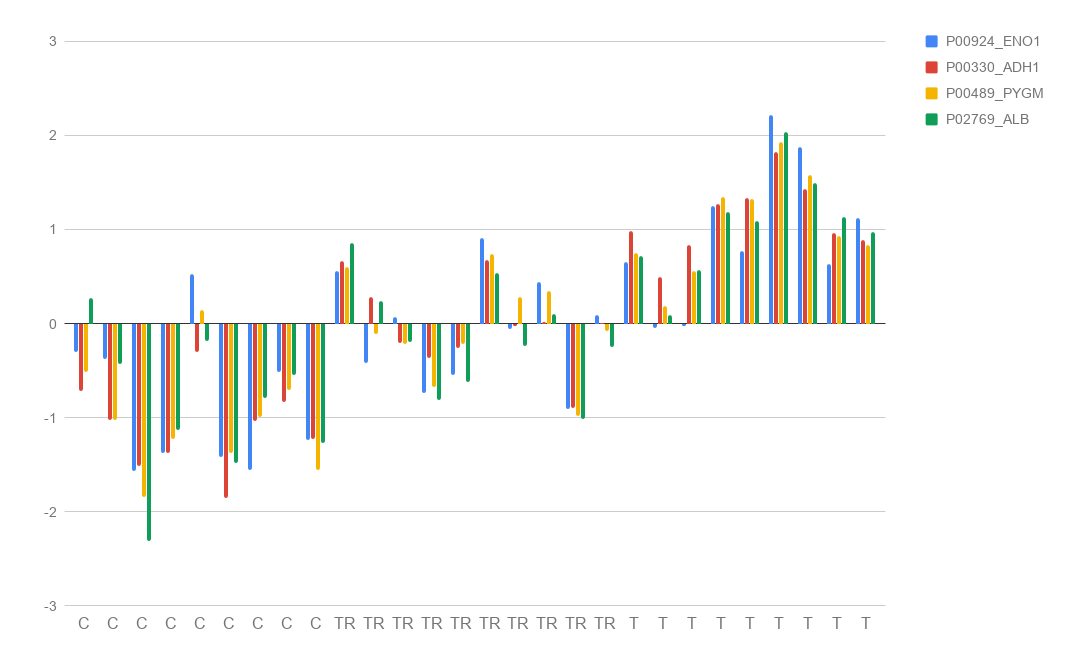
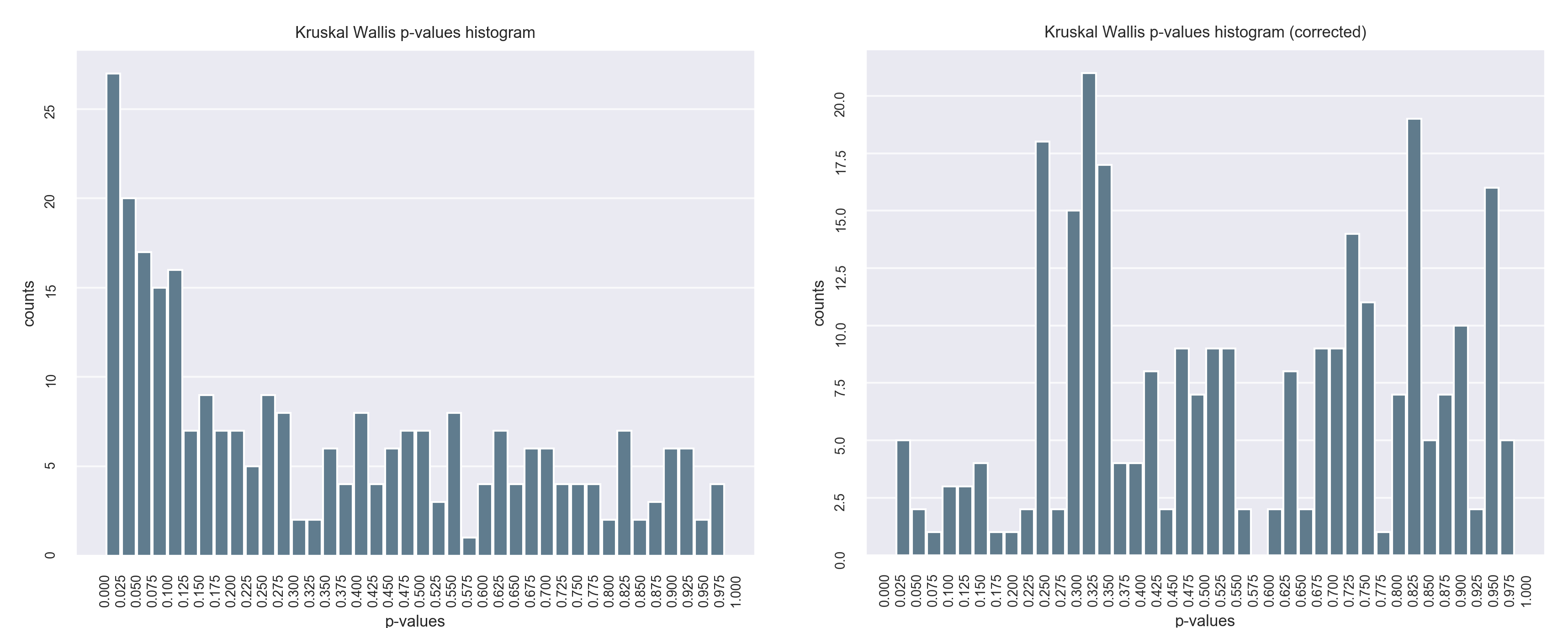
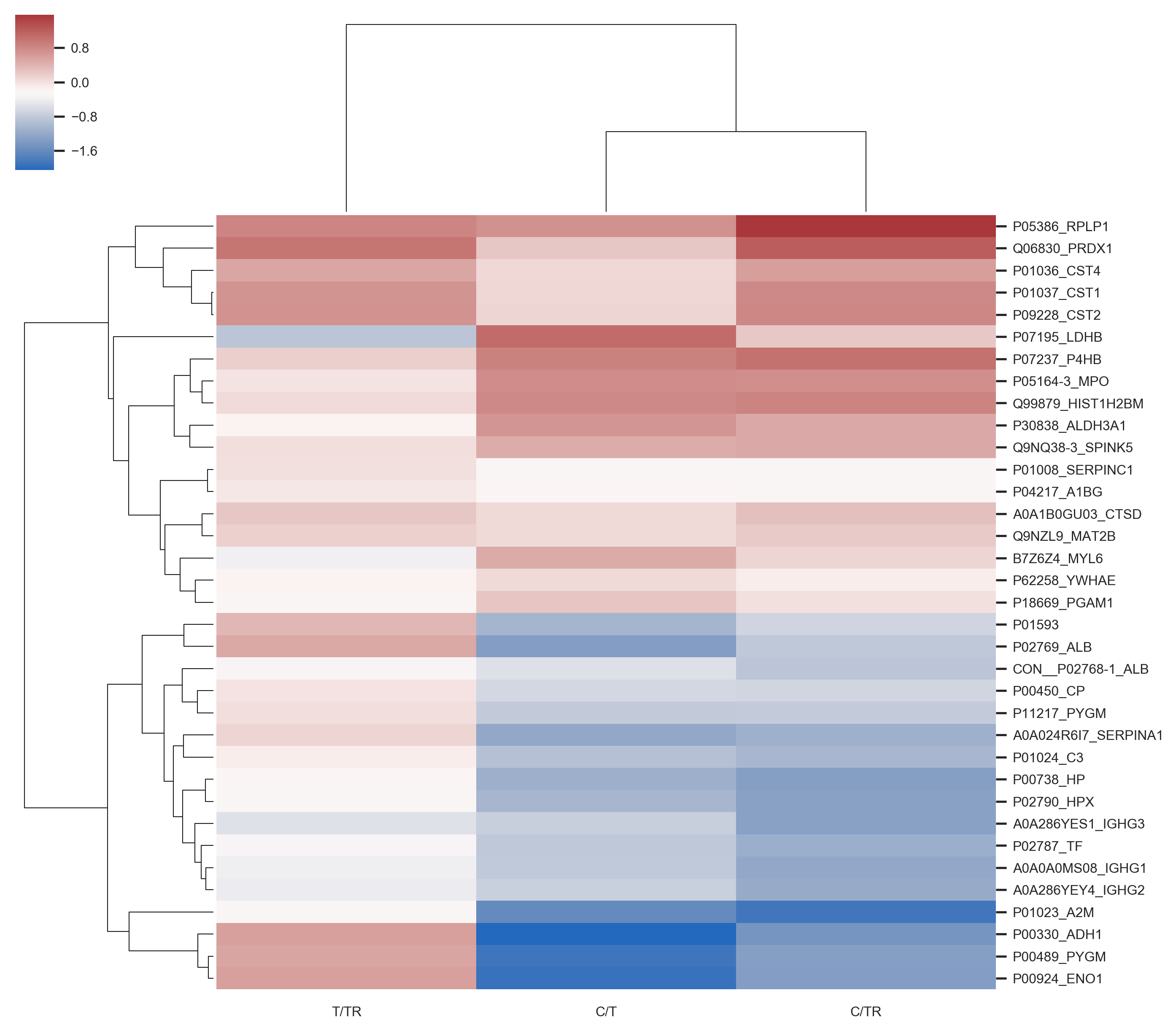
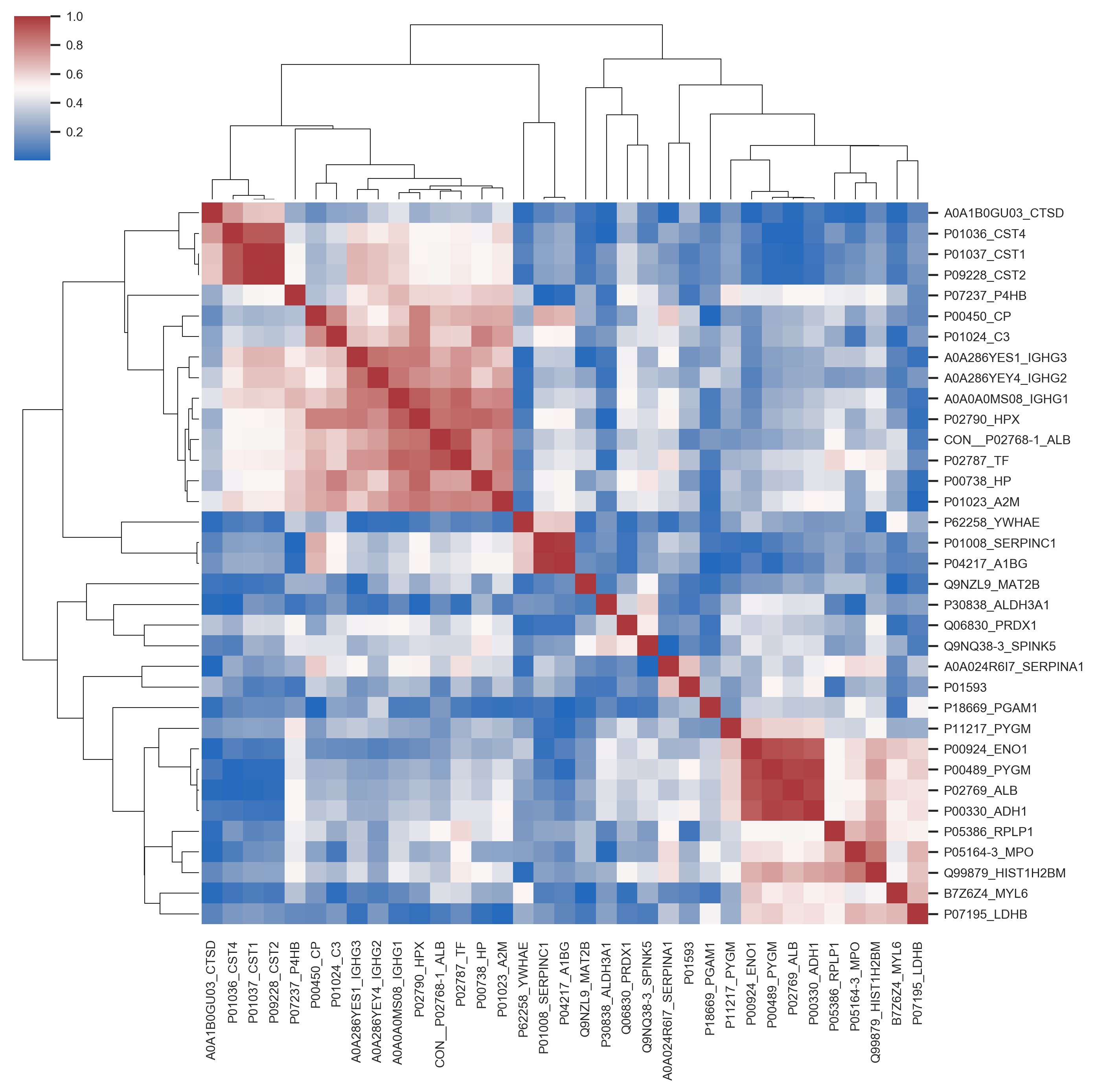
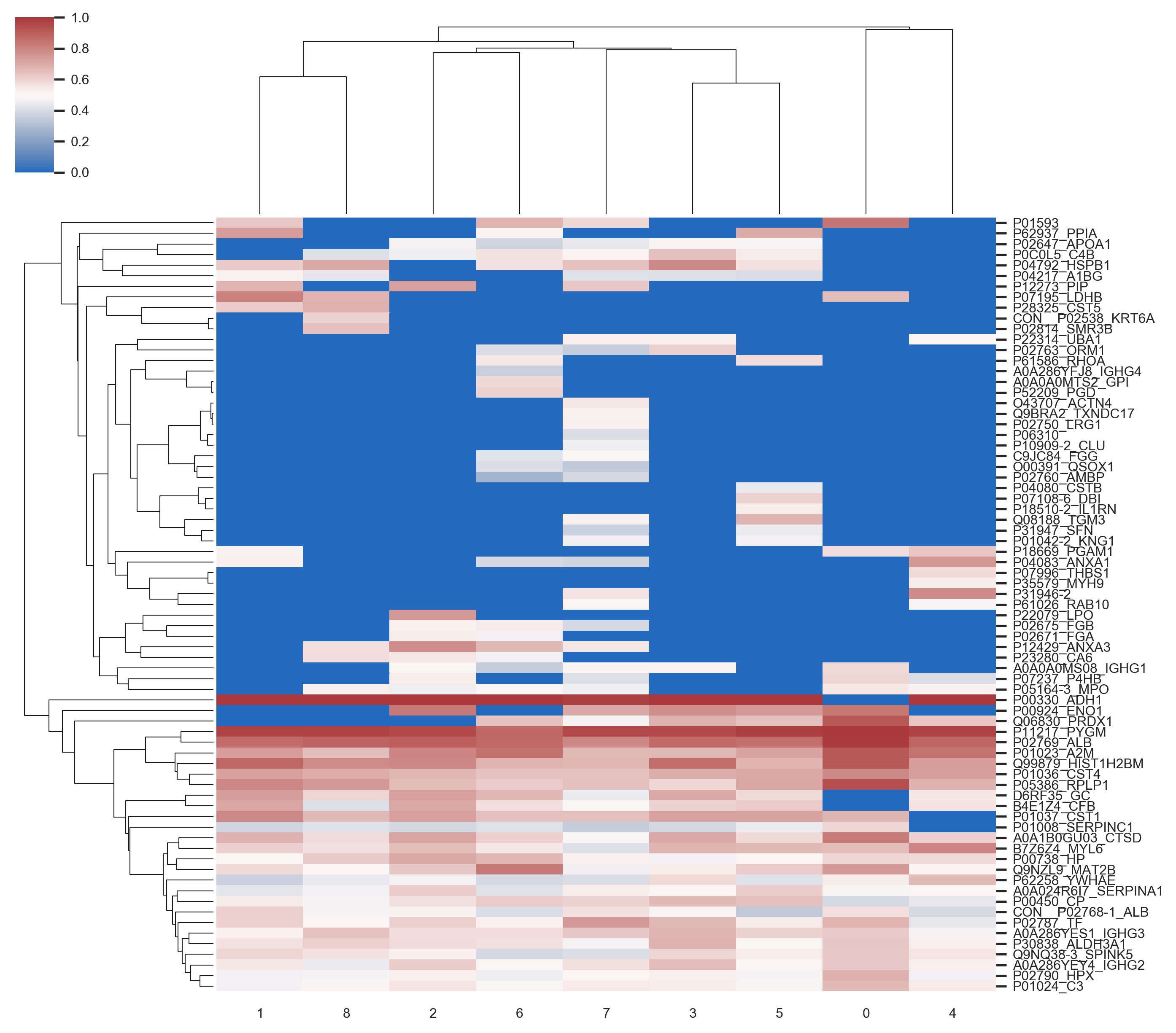
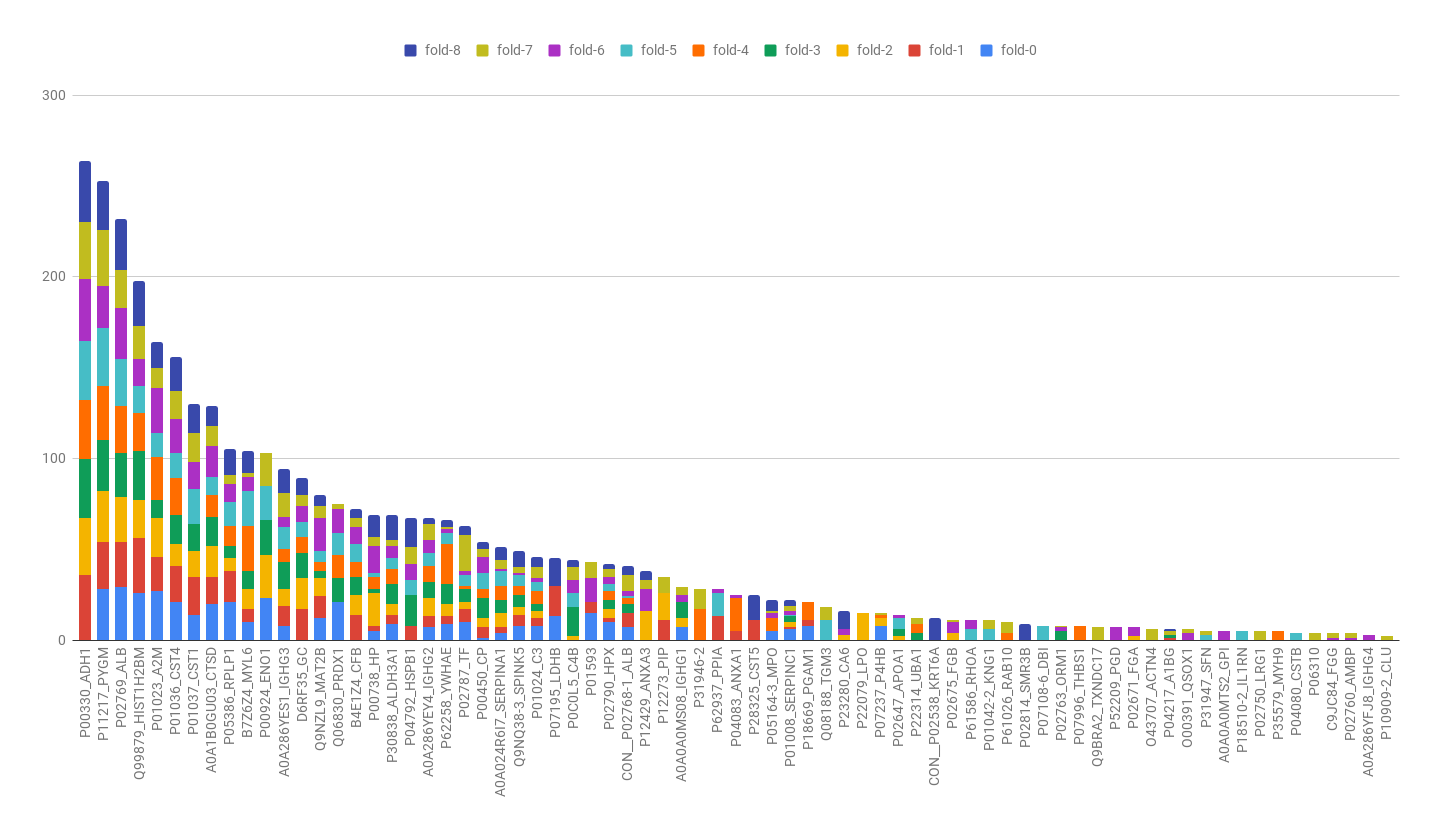
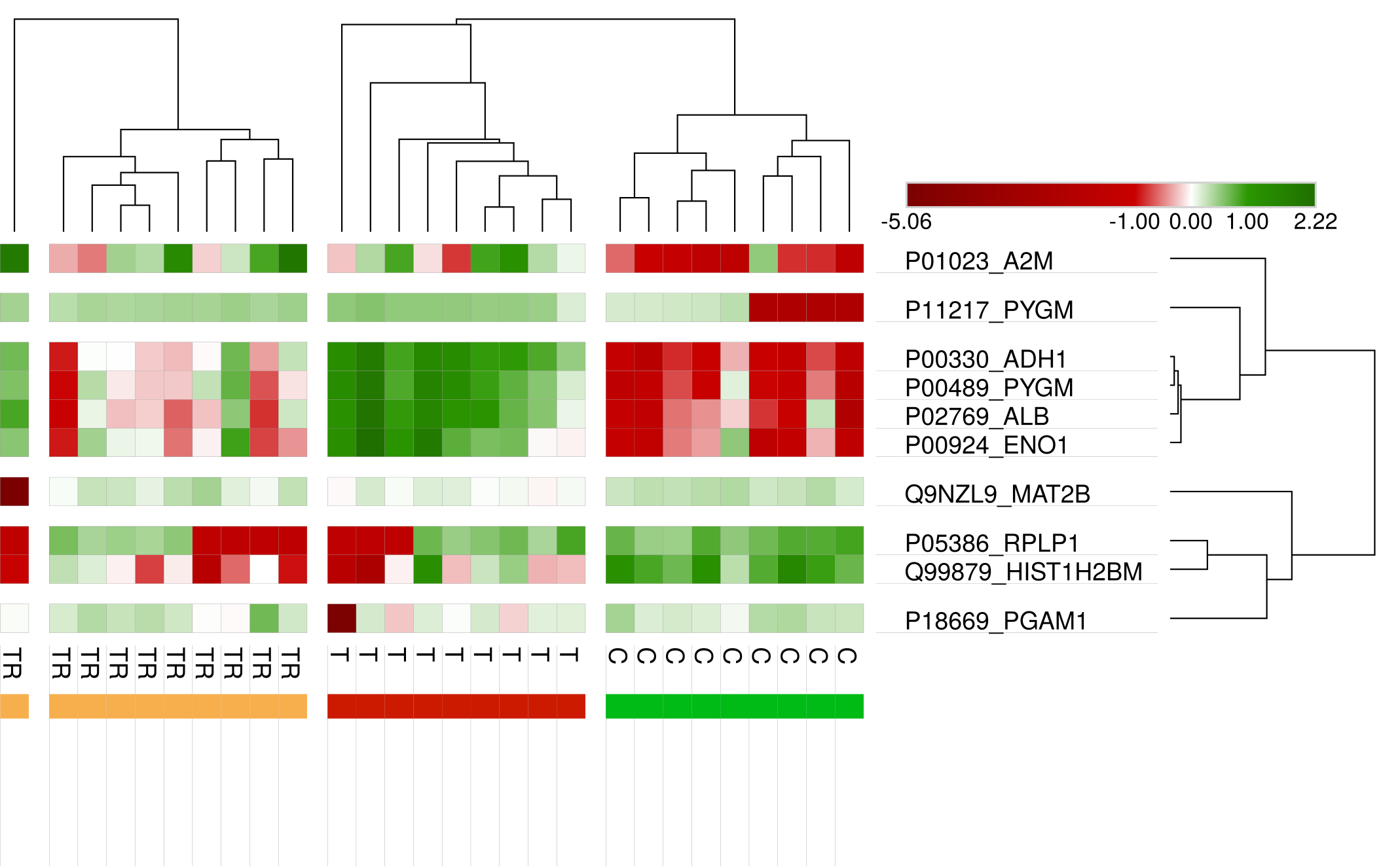
In this project I proposed the use of frequency of features (proteins) in good positions in ranks as an approach for feature selection in the context of Discovery Proteomics for Biomarkers research. These field usually have high-dimensional dataset with a much smaller number of samples.
I have used controlled samples in which proteins were added in different concentrations in different conditions: cancer, tumor removed and no-cancer. The biological experiments were developed by our collaborators from the Brazilian Bioscience National Laboratory. Proteins had their intensities quantified through Discovery Proteomics methods.
My method was capable of selecting the true markers and reveal information about instability of Discovery Proteomics and feature selection methods applied to this type of data. Visualization is of great importance for this category of work, once the experts are fundamental key and need to take decisions considering the high probability of bias and false discovery rate caused by the contrast between the number of samples and the number of variables in the dataset.
All results and research done is reported in my Doctoral dissertation.