I developed sTVis during an internship at Dalhousie University (Halifax, Canada) under supervision of professor Dr. Robet Beiko. The web-system allows the user to explore a phylogenetic tree (supertree) by exploring the edges that connect the internal nodes. Each edge may represent shared genes and SPR moves.
Summary
Gallery

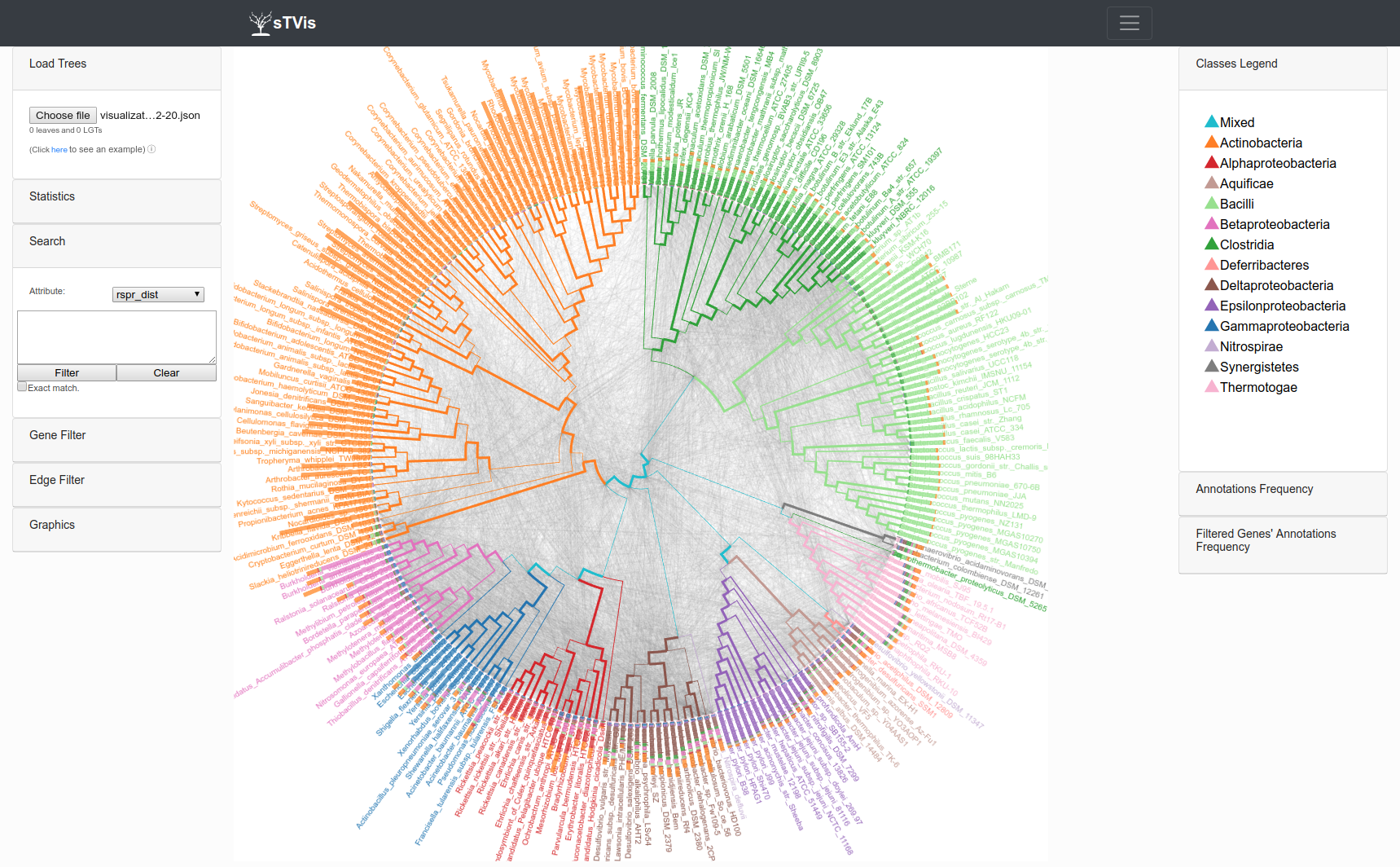
System overview
The visualization web-site.
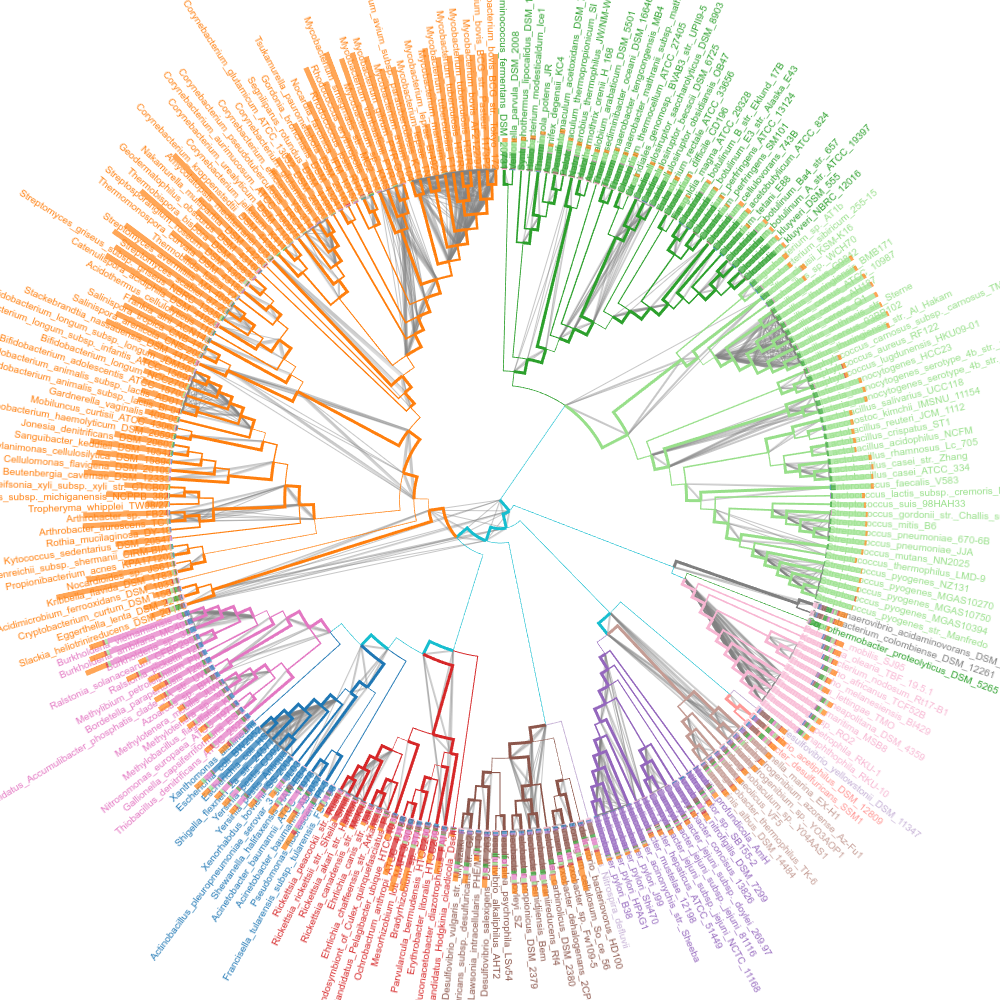
Short edges
Edges with small distance are shown.
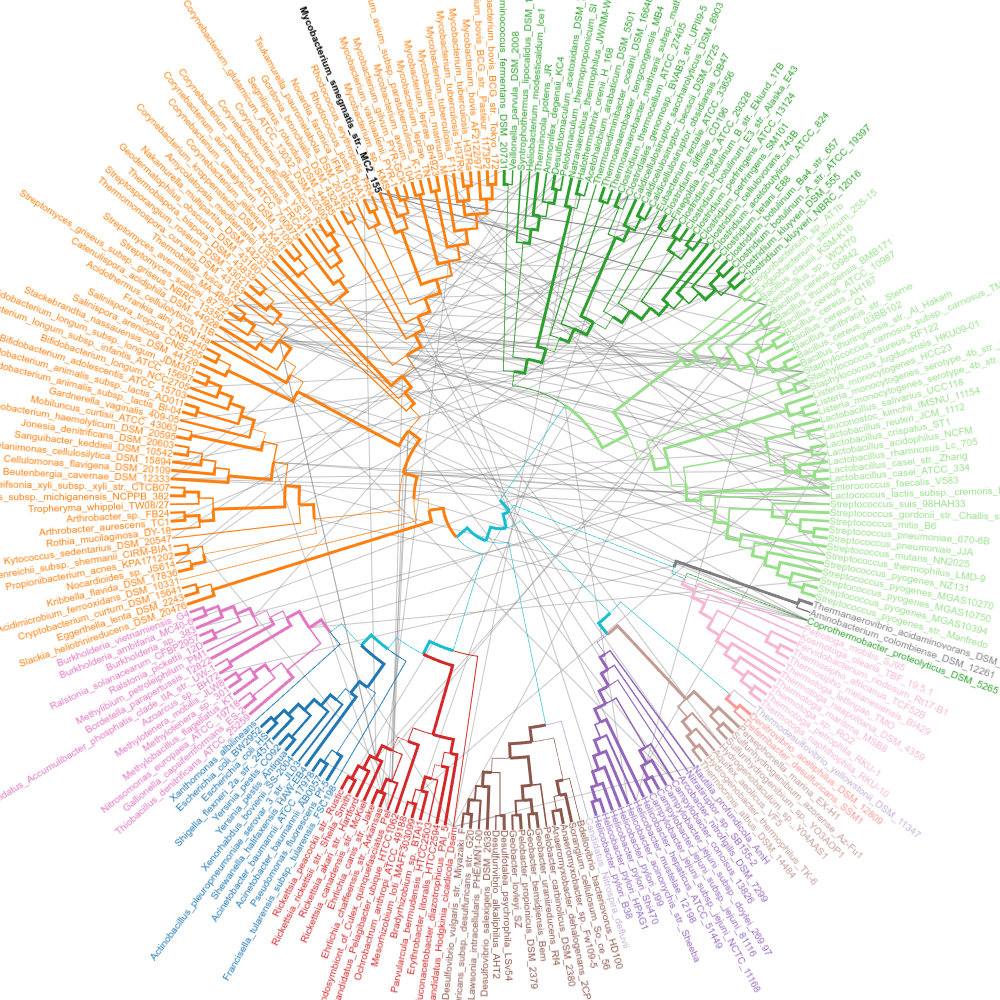
SPR edges
Long SPR edges.
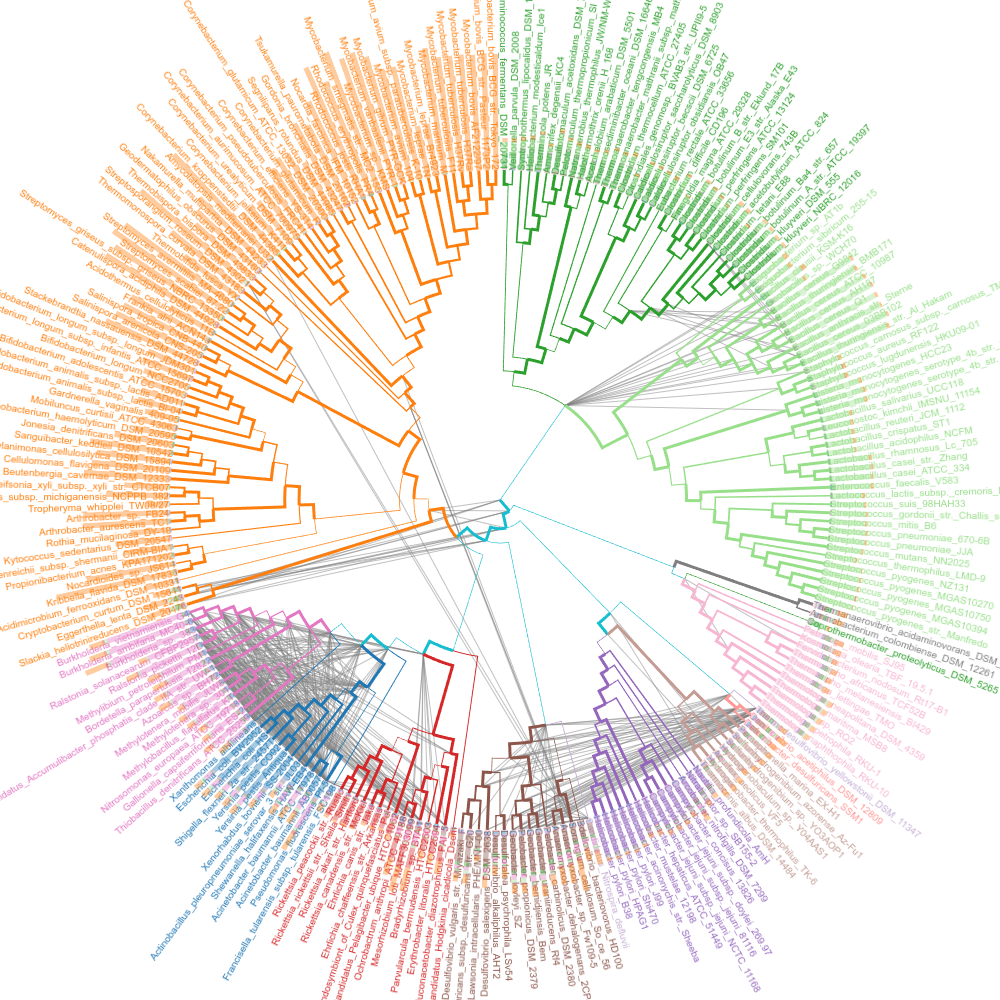
Shared genes
Interclass edges that represent more than 298 genes each. As expected, close classes have great number of genes in common.
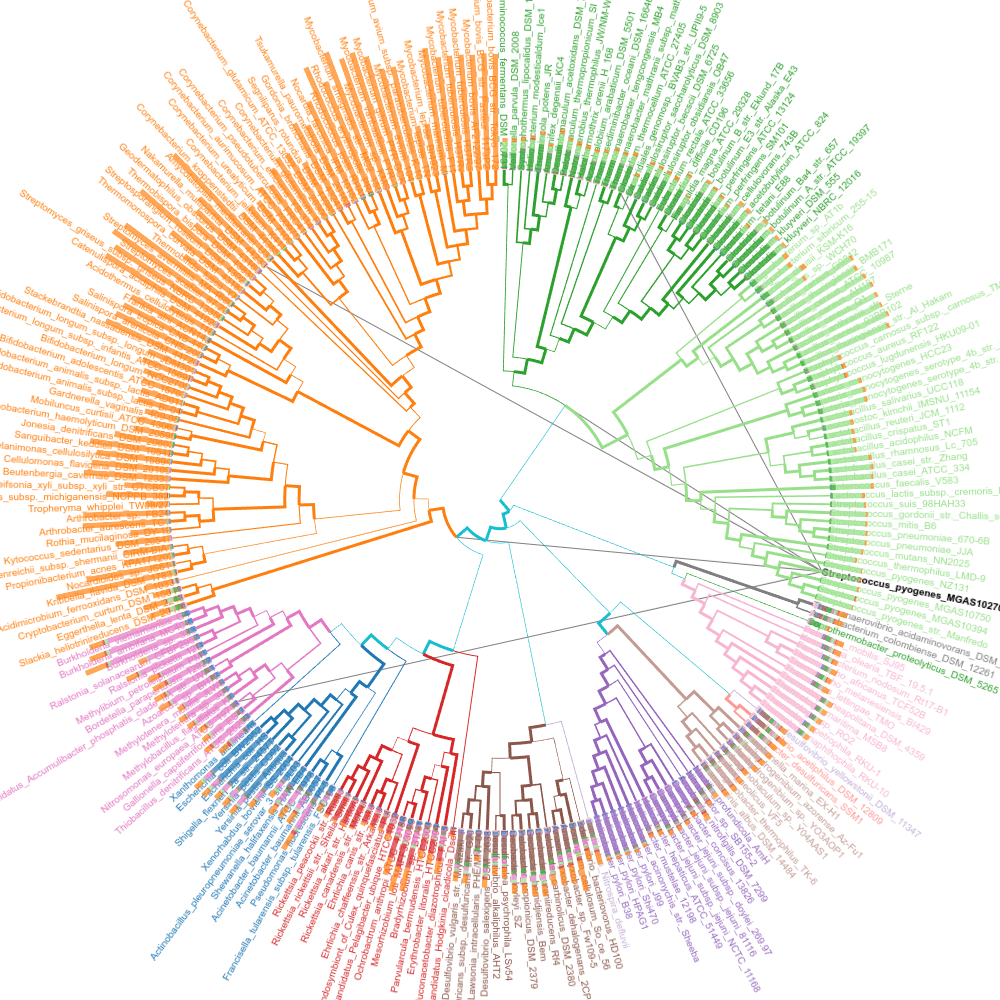
Streptococcus pyogenes
Edges that indicate shared genes with Streptococcus pyogenes.
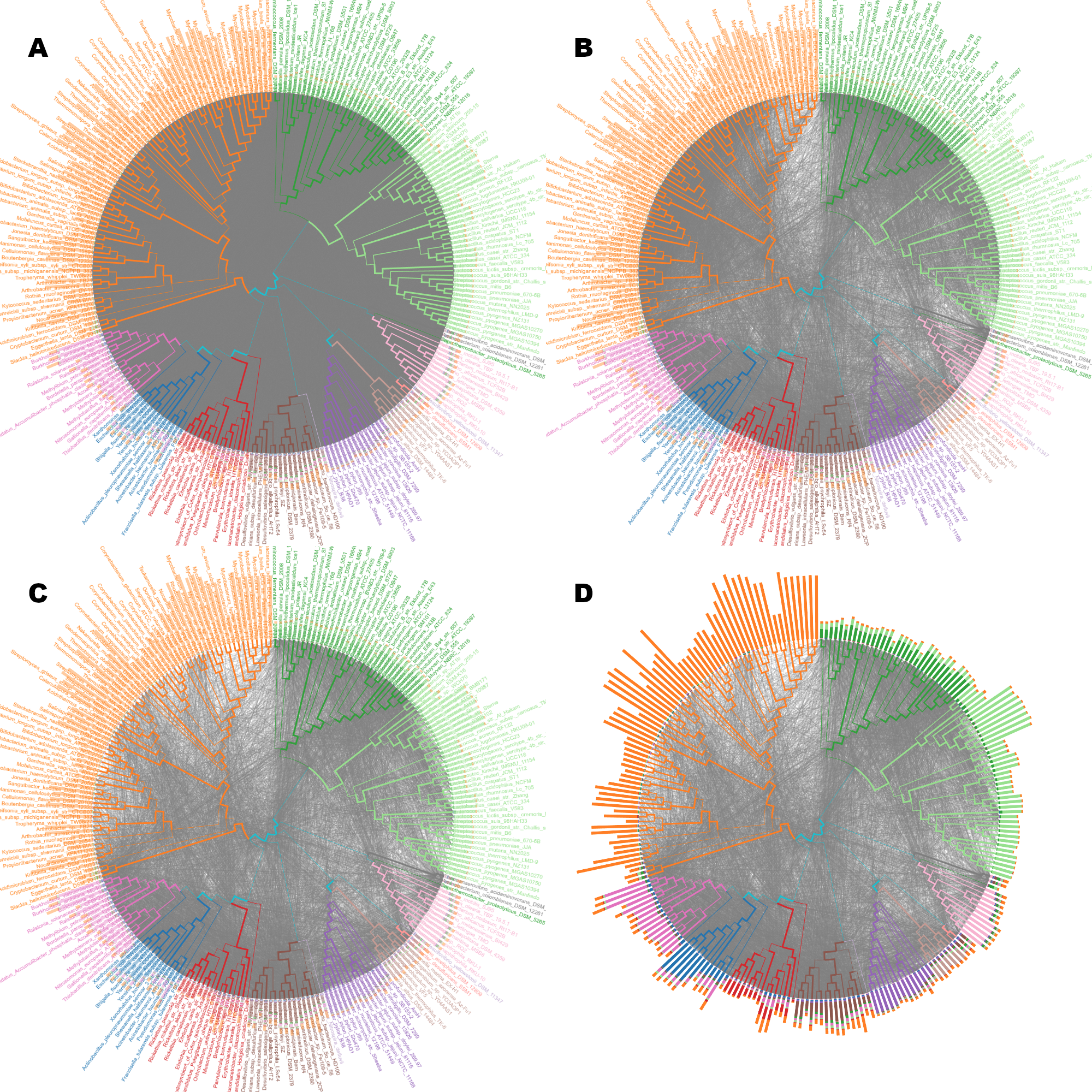
Sample similarities
Considering all features (A) and selected features (B, C, D).
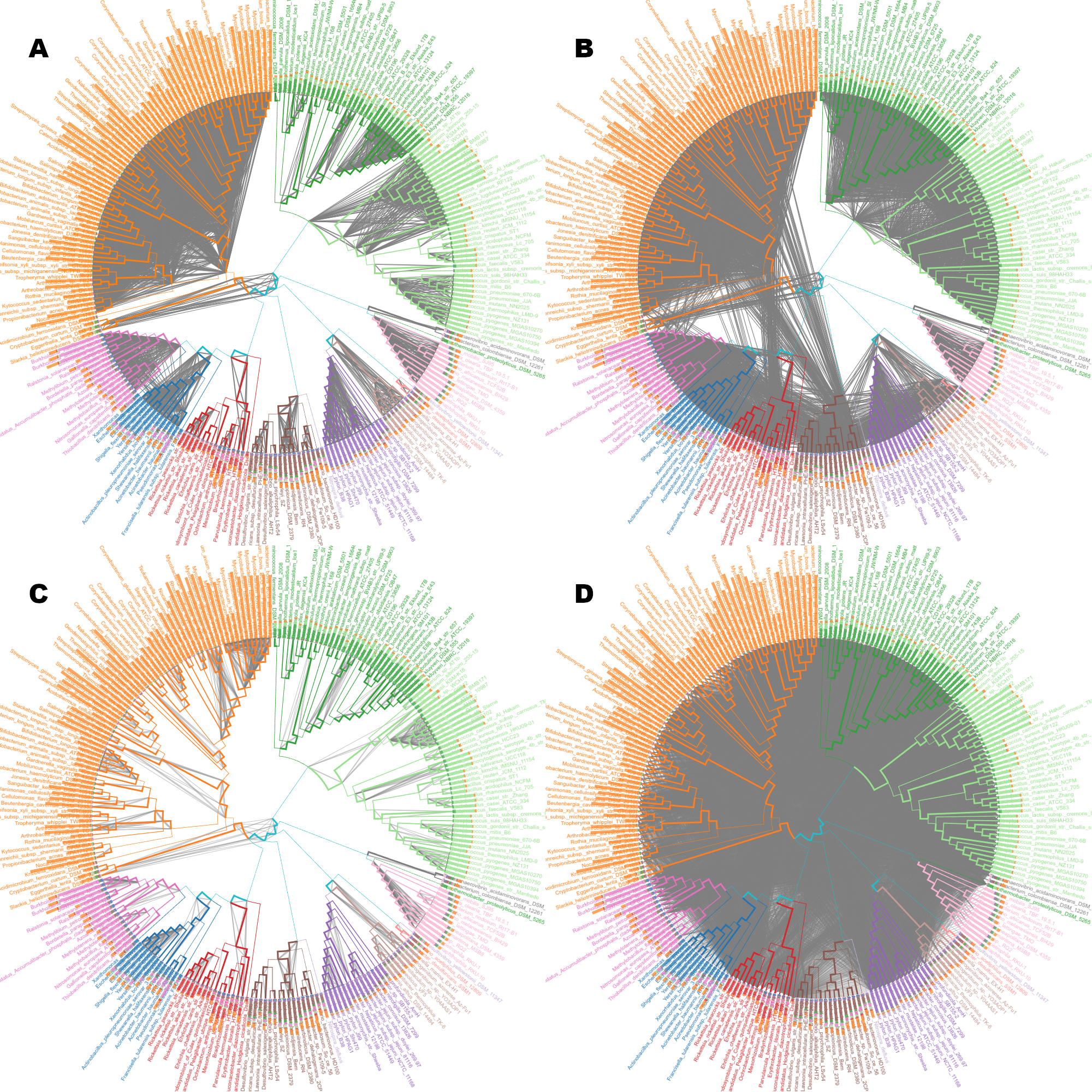
Shared genes
Example showing the power of the tool to show hundreds of thousands of edges. Filtering is applied on demand.
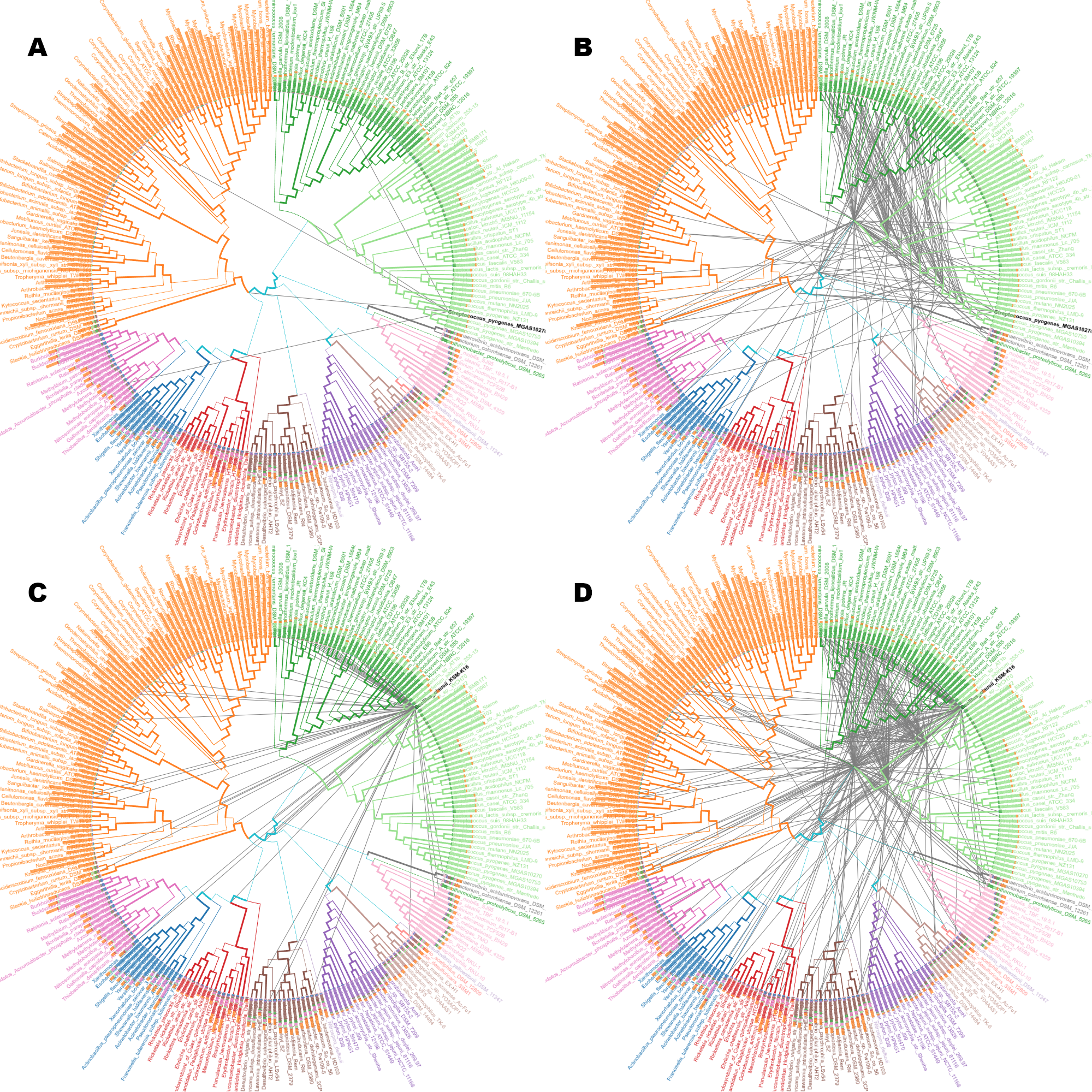
SPR moves
Comparison of SPR edges, fildering by distance/length.
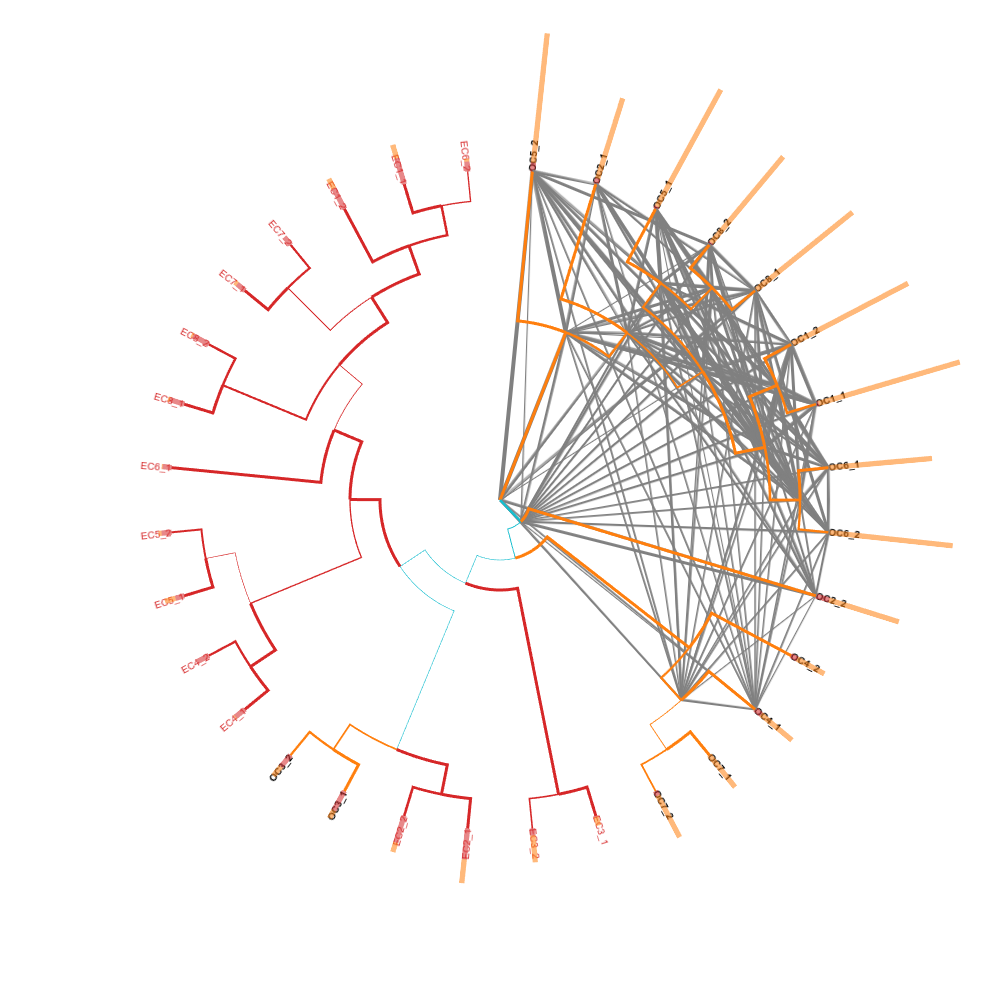
Proteomics supertree
Application of Supertree concept in showing relations between proteins of biological samples.
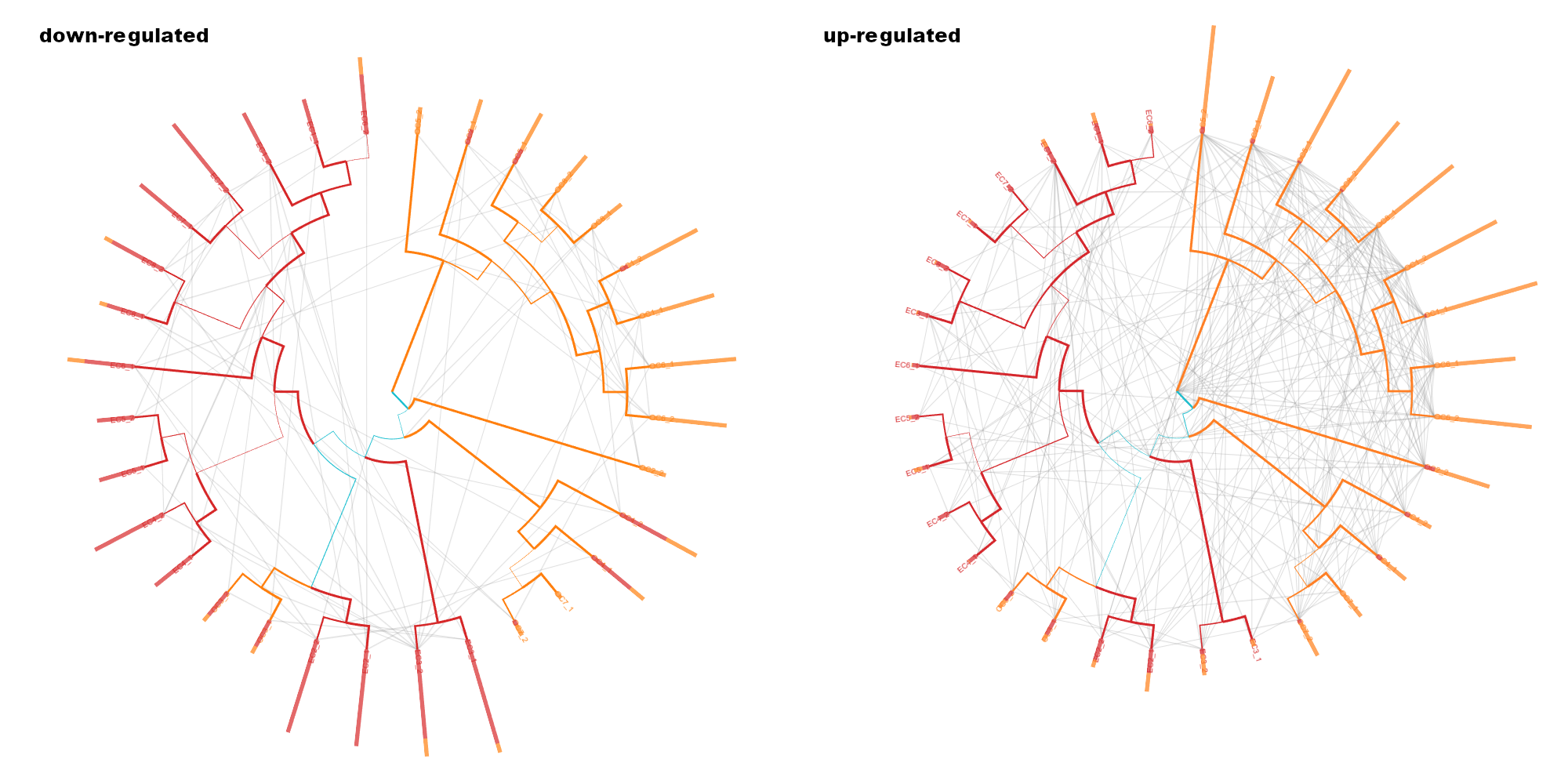
Proteomics supertree
Comparison of up and down regulated proteins as a supertree.